(going through this post again three years after I posted it. Made some, hopefully useful, changes)
(01.2018: further changes following DT comment)
Interaction are the funny interesting part of ecology, the most fun during data analysis is when you try to understand and to derive explanations from the estimated coefficients of your model. However you do need to know what is behind these estimate, there is a mathematical foundation between them that you need to be aware of before being able to derive explanations.
I plan to make two post on this issue, this first one will deal with interpreting interactions coefficients from classical linear models, a second one (which never came out) will look at the F-ratios of these coefficients and what they mean. I will only look at two-way interaction because above this my brain start to collapse. Some later post might be taking into account the extensive literature on these issues that I only started to scratch.
If you want to have a look at a clean page with code/figures go there: http://rpubs.com/hughes/15353
This post is divided in three parts: i) interaction between two categorical variables, ii) interaction between one continuous and one categorical variables and finally iii) interaction between two continuous variables.
Interaction between two categorical variables:
Let’s make an hypothetical examples: say we measured the shoot length (in cm) of some plant species under two different treatments: a temperature treatment with 2 levels (Control and Elevated temperature) and a nutrient treatment with three levels of nitrogen (Control, Medium, High nitrogen concentration). Following the recommendation of our stats professor we made a fully-factorial design and aim at look at the effect of these two treatments and their interactions on the shoot length.
#interpreting interaction coefficients from lm
#first case two categorical variables
set.seed(12)
f1<-gl(n=2,k=30,labels=c("Low","High"))
f2<-as.factor(rep(c("A","B","C"),times=20))
modmat<-model.matrix(~f1*f2,data.frame(f1=f1,f2=f2))
coeff<-c(1,3,-2,-4,1,-1.2)
y<-rnorm(n=60,mean=modmat%*%coeff,sd=0.1)
dat<-data.frame(y=y,f1=f1,f2=f2) summary(lm(y~f1*f2)) #output #Call: #lm(formula = y ~ f1 * f2) #Residuals: # Min 1Q Median 3Q Max #-0.199479 -0.063752 -0.001089 0.058162 0.222229 #Coefficients: # Estimate Std. Error t value Pr(>|t|)
#(Intercept) 0.97849 0.02865 34.15 <2e-16 ***
#f1High 3.00306 0.04052 74.11 <2e-16 ***
#f2B -1.97878 0.04052 -48.83 <2e-16 ***
#f2C -4.00206 0.04052 -98.77 <2e-16 ***
#f1High:f2B 0.98924 0.05731 17.26 <2e-16 ***
#f1High:f2C -1.16620 0.05731 -20.35 <2e-16 ***
#---
#Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
#Residual standard error: 0.09061 on 54 degrees of freedom
#Multiple R-squared: 0.9988, Adjusted R-squared: 0.9987
#F-statistic: 8785 on 5 and 54 DF, p-value: < 2.2e-16
Let’s go through each coefficient:
- (Intercept): plant shoots are predicted to have a length of 0.98cm under control temperature and nitrogen conditions, this is our baseline.
- f1High: plant shoots grow 3cm bigger under elevated temperature and control nitrogen conditions compared to the baseline
- f2B: plant shoots grow ~2cm smaller under medium nitrogen and control temperature condition compared to the baseline
- f2C: plant shoots grow 4cm smaller under high nitrogen and control temperature condition compared to the baseline
- f1High:f2B : effect of medium nitrogen condition in plant shoots under elevated temperature increase by approx. 1 compared to the effect of medium nitrogen condition under control temperature conditions. In other words, when the effect of adding medium nitrogen was to decrease plant shoots length by ~2cm in control temperature condition, in elevated temperature conditions adding medium level of nitrogen lead to shoot length of 0.97 -1.97 + 3 + 0.98 = 3 on shoot length.
- f1High: f2C : same as above for high nitrogen condition.
The best way to grasp what these means is still to make a quick graph:
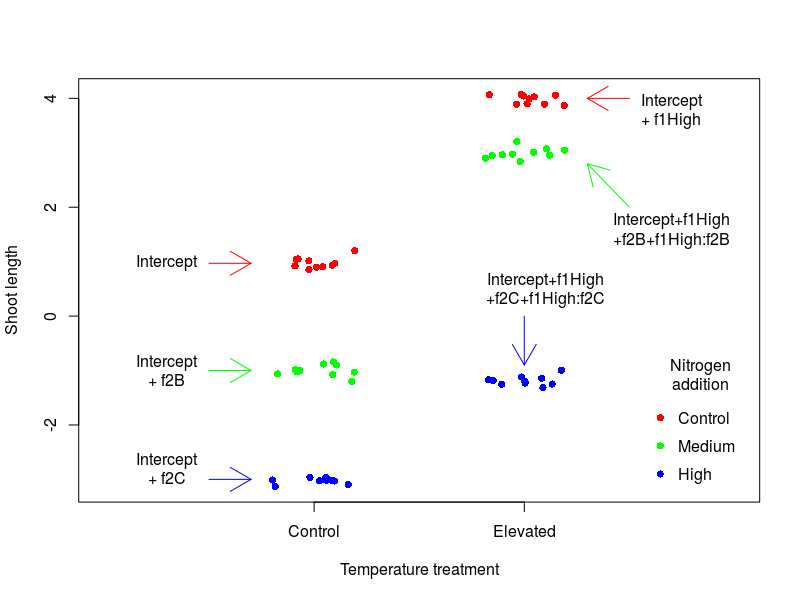
Not as easy to understand as one would thought …
Interaction between one continuous and one categorical variables
Say that we are weighting soil organisms in a field experiment where we added a temperature treatment with two levels (Control, Elevated) and we measured the soil nitrogen concentration (continuous variable measured in say mg per gram of soil), we would like to see the effects of the nitrogen concentration and its interaction with temperature on the biomass of soil organisms.
#second case one categorical and one continuous variable
x<-runif(50,0,10)
f1<-gl(n=2,k=25,labels=c("Low","High"))
modmat<-model.matrix(~x*f1,data.frame(f1=f1,x=x))
coeff<-c(1,3,-2,1.5)
y<-rnorm(n=50,mean=modmat%*%coeff,sd=0.5)
dat<-data.frame(y=y,f1=f1,x=x) summary(lm(y~x*f1)) #ouput #Call: #lm(formula = y ~ x * f1) # #Residuals: # Min 1Q Median 3Q Max #-0.9627 -0.2945 -0.1238 0.3386 0.9835 #Coefficients: # Estimate Std. Error t value Pr(>|t|)
#(Intercept) 1.35817 0.17959 7.563 1.31e-09 ***
#x 2.95059 0.03187 92.577 < 2e-16 ***
#f1High -2.63301 0.25544 -10.308 1.54e-13 ***
#x:f1High 1.59598 0.04713 33.867 < 2e-16 ***
#---
#Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#Residual standard error: 0.4842 on 46 degrees of freedom
#Multiple R-squared: 0.9983, Adjusted R-squared: 0.9981
#F-statistic: 8792 on 3 and 46 DF, p-value: < 2.2e-16
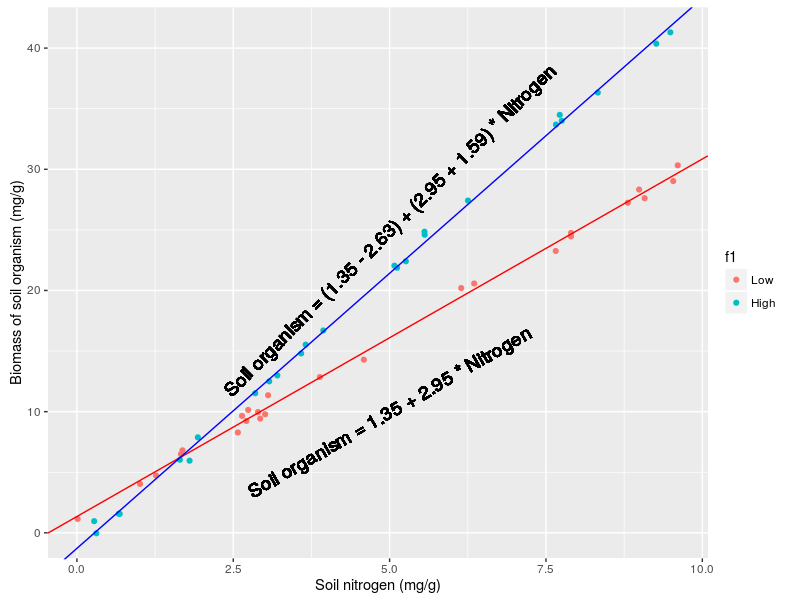
Again let’s go through the coefficients:
- (Intercept): Under control temperature condition and with a nitrogen concentration of 0 mg/g, soil organism biomass is 1.36 mg/g
- x: Adding 1mg/g of nitrogen increase soil organism biomass by 2.95 mg/g in control temperature conditions
- f1High: Under elevated temperature condition and for a nitrogen concentration of 0 mg/g soil organism biomass is 2.63 mg/g lower compared to control temperature condition
- x:f1High : the effect of soil nitrogen on soil organism biomass is higher by 1.59 under elevated temperature condition compared to control condition. In other words the slope between soil nitrogen and soil organism biomass is 2.95 + 1.59 = 4.54 in elevated temperature condition.
And a graph to grasp:
Interaction between two continuous variables
Now for the last possible case imagine that we wanted to compare our experimental results to what is happening in the “real” world. We went happily outside measured air temperature at the soil surface and collected soil samples from which we got soil nitrogen concentration (in mg per g of soil) and soil organism biomass (in mg per g of soil). Again we build a linear model to test the effect of temperature and nitrogen plus their interaction.
# third case interaction between two continuous variables
dat <- data.frame(nitrogen = runif(50, 0, 10),
temperature= rnorm(50, 10, 3));
modmat <- model.matrix(~ nitrogen * temperature, dat);
coeff <- c(1, 2, -1, 1.5);
dat$soil <- rnorm(50, mean = modmat %*% coeff, sd = 0.5);
#fit a model
m <- lm(soil ~ nitrogen * temperature, dat);
summary(m)
Call:
lm(formula = soil ~ nitrogen * temperature, data = dat)
Residuals:
Min 1Q Median 3Q Max
-0.78742 -0.21694 0.00031 0.26940 0.92838
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.541509 0.516898 2.982 0.00456 **
nitrogen 1.930706 0.083030 23.253 < 2e-16 ***
temperature -1.046353 0.050237 -20.828 < 2e-16 ***
nitrogen:temperature 1.505170 0.008279 181.815 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.4326 on 46 degrees of freedom
Multiple R-squared: 0.9999, Adjusted R-squared: 0.9999
F-statistic: 2.368e+05 on 3 and 46 DF, p-value: < 2.2e-16
The coefficient means:
- (Intercept): at 0°C and with nitrogen concentration of 0 mg/g, soil organism biomass is 0.96 mg/g
- nitrogen: increasing nitrogen concentration by 1 mg/g at a temperature of 0°C increase soil organism biomass by 1.93 mg/g
- temperature: increasing temperature by 1°C at a nitrogen concentration of 0 mg/g reduce the soil organism biomass by 1 mg/g
- nitrogen:temperature : (sit down and breath deeply) the effect of nitrogen on soil organism biomass increase by 1.50 for every unit increase in temperature. For example at a temperature of 0°C the slope between soil organism biomass and nitrogen is 1.93, at a temperature of 1°C this slope becomes: 1.93 + 1.50 * 1 = 3.43. This also works the other way round (ie effect of temperature on soil organism biomass become more positive as nitrogen concentration increase).
And a final graph:
new_d <- expand.grid(nitrogen=0:10,temperature=c(3.55,10,16)) new_d$Pred <- predict(m,new_d) ggplot(dat,aes(x=nitrogen,y=soil,color=temperature))+ geom_point()+ geom_line(data=new_d,aes(group=temperature)) + scale_color_continuous(low="green",high="red") + geom_text(aes(x=5,y=20,label="Slope: (1.93 + 1.50 * 3.55) * nitrogen"),color="green",angle=13,family="Arial") + geom_text(aes(x=5,y=65,label="Slope: (1.93 + 1.50 * 10) * nitrogen"),color="brown",angle=30,family="Arial") + geom_text(aes(x=5,y=130,label="Slope: (1.93 + 1.50 * 16) * nitrogen"),color="red",angle=45,family="Arial")
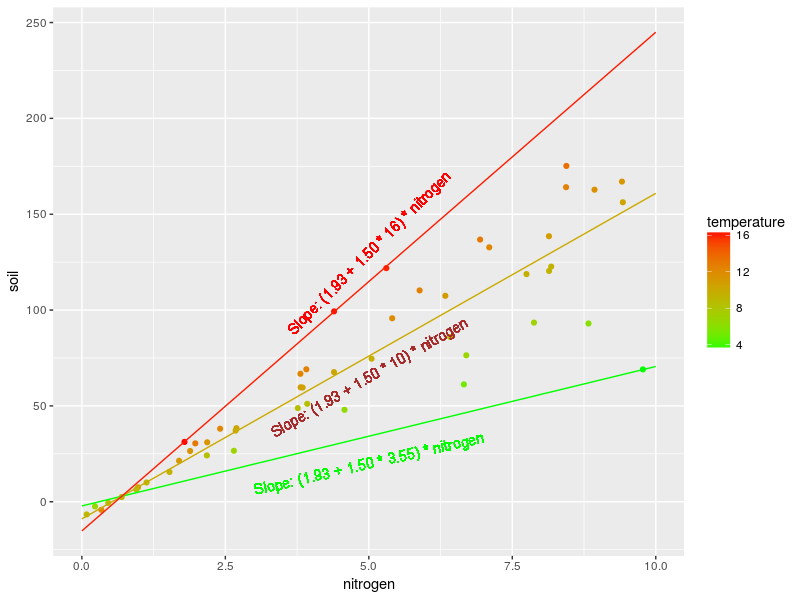
In this graph we drew the regression line of the relation between soil organism and soil nitrogen at three different temperature: at 3.55, 10 and 16°C. The text represent the slopes between soil organism and nitrogen and these different temperature. Note that you need to add the intercept plus the main effect of temperature to get the predicted value. For instance at nitrogen 5 mg/g and temperature 10°C we have:
soil organism = 1.54 – 1.04 * 10 + (1.93 + 1.50 * 10) * 5 = 75.79
So next time you want to fit 5-way interactions try to remember that already understanding two-way interaction need some thinking …
Happy coding.

Thanks for this post, it helped me a lot. I’ve been trying to find a practical explanation for interactions in linear models for ages now.
I’m fairly new to this study, so it would have helped if you explained how to interpret the plots with few extra words. To me they weren’t self-explanatory at all even though I study mathematics. The explanations on the lm results were also little vague, it took me some time to fully digest those.
Thanks for your comments. Went through the post a second time and tried to improve readability, let me know if you have further suggestions!
Thanks for this! One question: In the interaction between continuous variables section, why is the intercept in the graph 0.63 when the intercept in the linear model is 0.78?
Thanks Keaton for your kind words, I wrote the code “on-the fly” without setting seeds and as I re-wrote the article I did not update the last figure, the two should now match!
Good post, there is an error though in the first section. The intercepts are correct in your graph, but not so in your explanation. You write -1.97 + 0.98 = – 0.99 on shoot length for med N and high temp. You forgot to add the high temp deviation here. Correct answer, as shown in graph is 0.98+3+0.99-1.97 = 3. Same for •f1High: f2C.
Cheers
Thanks for spotting this mistake, finally took the time to change the text 🙂
Thanks a lot, very clear .
Considering your last plot, with three different slopes for three different temperature levels. According to the formulas associated with the graphs, the predicted soil organism levels seem to only depend on Nitrogen and Nitrogen:Temperature. Nevertheless, we have estimated a significant baseline temperature slope. Why is this missing in the formulas? For instance, for Temperature = 5, I would expect the formula to contain + 1.995*5 as well, but it doesn’t. Same goes for Temperature = 10.
Best
Mathias
Hello. Thanks for the post. I am also confused about the third example. Assuming that the intended equation is:
Y = α + β1 X1 + β2 X2 + β3 ( X1 * X2) + ε
which in reduced form is this:
Y = α + β1X1 + ( β2 + β3 * X1) * X2 + ε
Which, I think, is what you are using in your plots. Isn’t it the case that your plots are missing β1X1 as the commenter above suggested?
Also, plugging in values (X1 = 5, X2 = 17) to this equation produces:
Soil = .96 + 1.995 * 5 + ( -0.993288 + 1.499595 * 5) * 17 + ε
and thus:
Soil = 121.51
Which doesn’t seem to match the values on your plot, even if I omit the β1X1 term as you did in my suggested change to the equation.
In fact, when I extract values from your model using effect() your plot values still do not seem to line up, though the result from my suggested equation does (or is close given that error was not included perhaps?):
x1*x2 effect
x2
x1 2.4 6 9.6 13 17
0.07 -0.9309848 -4.129814 -7.328644 -10.34976 -13.90402
2 9.9031351 17.098106 24.293077 31.08833 39.08274
5 26.7437361 50.094873 73.446010 95.49986 121.44557
7 37.9708033 72.092718 106.214633 138.44089 176.35412
10 54.8114043 105.089485 155.367566 202.85242 258.71696
Granted, I am very new to this and do not really understand the intricacies of these models. But I would appreciate it if you could comment back with a response.
Thanks!
Thanks DT for your comment, I went through he article again and re-wrote the last section as it was confusingly written between x1 and nitrogen and x2 and temperature. I hope that it is know clearer. Please have a look through it and let me know if some things are still unclear.
Concerning your comments, in the graph I now write down the equation of the slopes, so not taking into account the intercept and the main effect of temperature (which is constant along the drawn lines).
So I would write the full equation:
Soil = a + b_1 * nitrogen + b_2 * temperature + b_3 * nitrogen * temperature
into
Soil = a + b_2 * temperature + (b_1 + b_3 * temperature) * nitrogen
The last term in this equation represent the written down slopes, the first two terms being constant once we set the temperature to a certain value.
Also note that I use the predicted value from the model, so not taking into account residual deviation (the epsilon term in your equation).
Thanks again for your comment and your attention to important details.
L
You explanation about the output of the interaction between two categorical variables was very helpful as well as instructions on how to graph it. Lets say that you added a random effect like plots where you collected your samples. The output is the same plus this:
Random effects:
Groups Name Variance Std.Dev.
plots (Intercept) 0.3007 0.5484
Do you need to add the variance or st.dev to the equation when you graph it? Or what would this mean in your elevation-nitrogen example?
Hi Yvan, this is a great question because this deserve a post in itself, thanks for giving me this idea! In a few words with a random effect you would need to take your fixed-effect coefficient (say its value is 2) and add to it the random terms either by taking the fitted values with something like: 2 + ranef(model)$plots, or by sampling new random effects with: 2 + rnorm(10,0,0.5484). Wait for my post for the full answer 🙂
Could you expand this post with a Interaction between two continuous variables and a categorical variable? Because I find it very tricky to interpret..
I am writing a blog post on three-way interactions that hopefully should allow you to see a bit clearer into this 🙂